Dr. Jędrzej Jakub Szymański
Research focus
Our group examines mechanistic relationships between genetic information and crop quality traits. Utilizing deep learning on extensive sequence and omics datasets, we identify and functionally characterize genetic sequence features that affect gene expression and translate to macro-phenotypes. Our strategy integrates multi-modal data, including genomic, transcriptomic, metabolomic, and phenomic information.
The five most important publications
- E. Korenblum, Y. Dong, J. Szymanski, S. Panda, A. Jozwiak, H. Massalha, S. Meir, I. Rogachev, A. Aharoni, Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proceedings of the National Academy of Sciences. 117, 3874–3883 (2020).
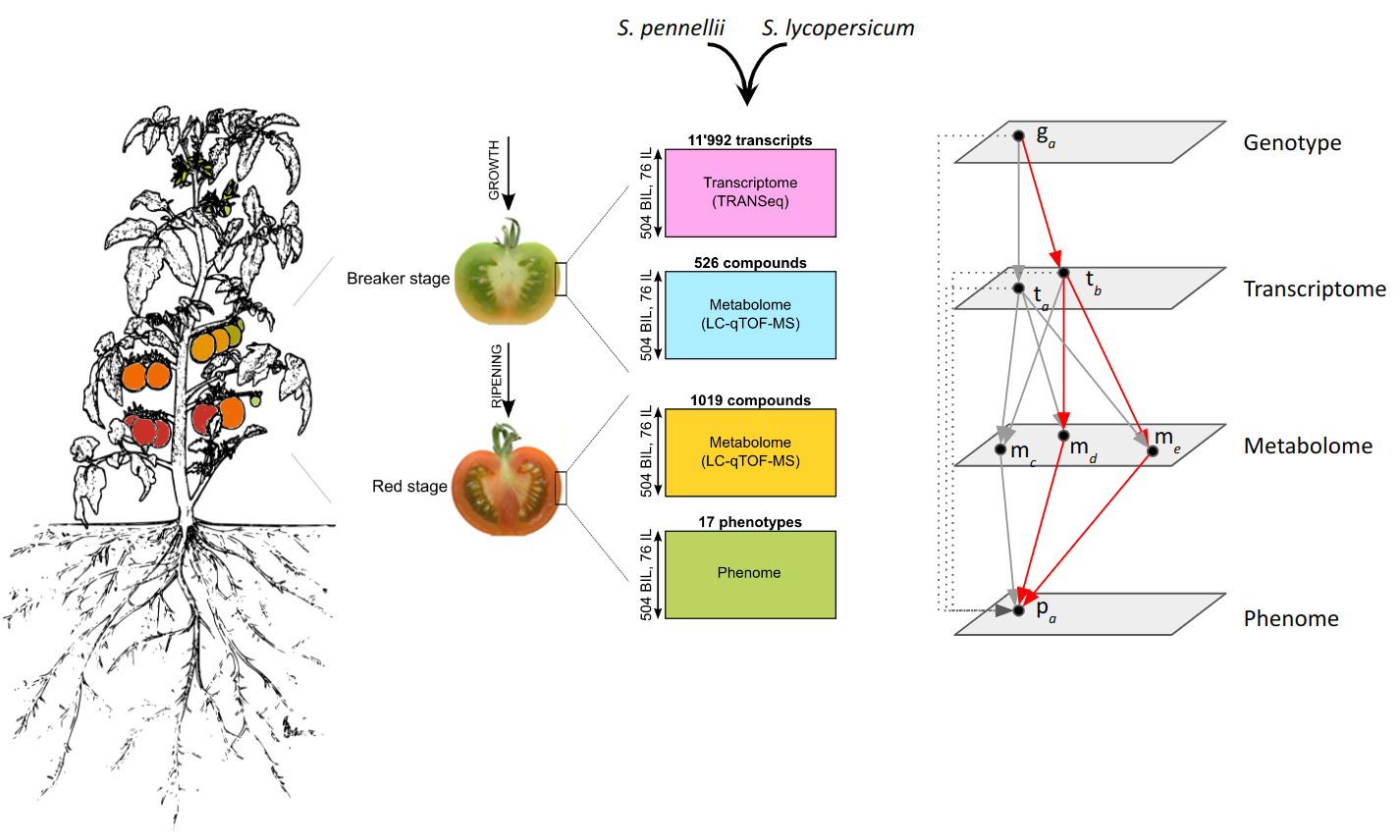
- J. Szymanski, S. Bocobza, S. Panda, P. Sonawane, P. D. Cárdenas, J. Lashbrooke, A. Kamble, N. Shahaf, S. Meir, A. Bovy, J. Beekwilder, Y. Tikunov, I. Romero de la Fuente, D. Zamir, I. Rogachev, A. Aharoni, Analysis of wild tomato introgression lines elucidates the genetic basis of transcriptome and metabolome variation underlying fruit traits and pathogen response. Nat. Genet.52, 1111–1121 (2020).
H. Redestig, J. Szymanski, M. Y. Hirai, J. Selbig, L. Willmitzer, Z. Nikoloski, K. Saito, Data integration, metabolic networks and systems biology. Annual Plant Reviews, 261–316 (2018).
J. Szymanski, Y. Levin, A. Savidor, D. Breitel, L. Chappell-Maor, U. Heinig, N. Töpfer, A. Aharoni, Label-free deep shotgun proteomics reveals protein dynamics during tomato fruit tissues development. Plant J.90, 396–417 (2017).
J. Szymanski, Y. Brotman, L. Willmitzer, Á. Cuadros-Inostroza, Linking gene expression and membrane lipid composition of Arabidopsis. Plant Cell. 26, 915–928 (2014).
Dr. Jędrzej Jakub Szymański

Institute of Bio- and Geosciences - Bioinformatics (IBG-4)
Forschungszentrum Jülich
RG Network Analysis and Modelling
Leibniz Institute of Plant Genetics and Crop Plant Research
https://www.ipk-gatersleben.de/en/research/molecular-genetics/network-analysis-and-modelling