How to follow corn genes into the past
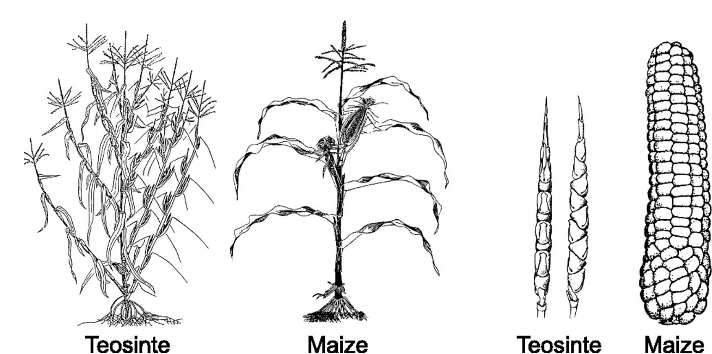
As one of the most important crops, maize is known worldwide and probably conjures up a similar image in most minds - a thickened spindle, densely studded with richly yellow colored seeds (maize kernels), which are tightly encased by leaves and with silky lustrous fibers at the top of the cob. This is the picture of modern, high-yield hybrid maize grown in most of the world today. However, if one looks at the ancestor of corn - teosinte, a wild-growing maize species concentrated primarily in Central America, clear differences in plant architecture and the number and shape of ears/cobs become clear at first glance (Fig. 1A). As an outcrossing species with a double set of chromosomes, the recurrent effects of recombination and the exertion of evolutionary forces during the domestication of teosinte, which began about 9000 years ago, led to gradual changes in the chromosome set that explain the presentday differences between teosinte and modern maize.
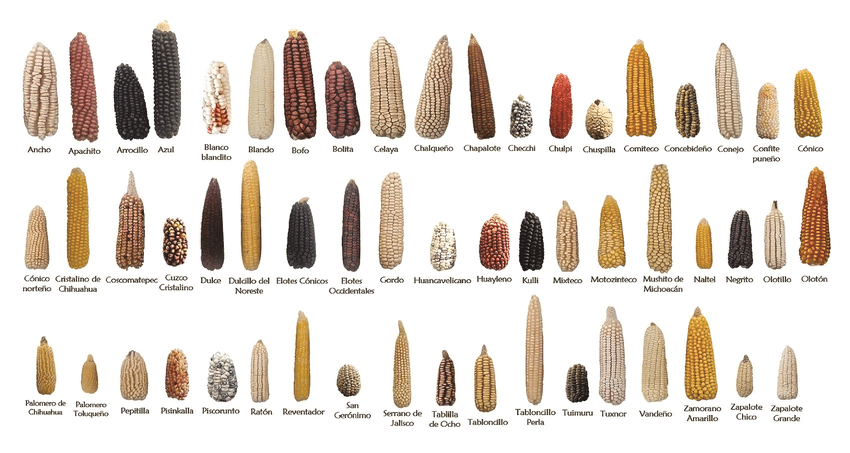
Today, modern cultivated maize is grown almost everywhere in the world, including almost everywhere in Europe. However, when it comes to corn today, there is still more diversity than one might think (Fig. 1B). This is because maize has a complicated history, characterized by a succession of demographic events, like population bottlenecks and expansions, migration and mixing between different regional populations, resulting in a variety of different regional maize populations with diverse gene pools. In Europe alone there are hundreds of local varieties, so-called landraces, which have been improved solely through traditional agricultural methods. These varieties were used extensively until the 1960s, when they were supplanted by the more productive, but also less diverse, modern maize.
Nowadays, these local landraces are mainly stored in gene banks. Many of these landraces are particularly well adapted to their locations and some of them have properties desired in plant breeding in the form of rare genetic variants, which have been lost in modern maize with its strong breeding focus on yield. The origins and history of these gene variants are still unclear today. When did they first appear? Were they already present in teosinte? How did they get to Europe? Or did they only appear when corn was first brought to Europe around 1492?
To find this out, we need to follow the maize gene variants back in time. The rationale behind this idea is based on the fundamental notion that the ancestral relationships of all maize individuals can theoretically be described by a single genealogy. Diploid sexually reproducing organisms, to which maize belongs, receive one set of chromosomes from their ”mother” and one from their ”father”, which in turn received one set of chromosomes from each of their parents, and so on. This type of inheritance of genetic material makes it possible to visualize the ancestral relationships between individuals as a family tree. This representation works both between individuals and between DNA sequences. However, a tree only accurately describes the ancestry of a DNA sequence if it is passed down through generations as a unit (haplotype). The rearrangement of genetic material (recombination) during gamete formation (meiosis) divides DNA segments and breaks up these units. As one traverses the genome, different parts of the genome have progressively different ancestors and must be described in terms of different trees (Fig. 2a). This results in a pedigree network that represents all ancestral relationships within a collection of chromosome sets from a large number of individuals (Fig. 2b).
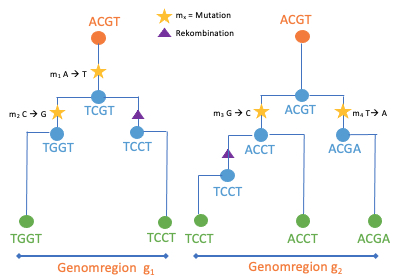
(a) Single tree representation
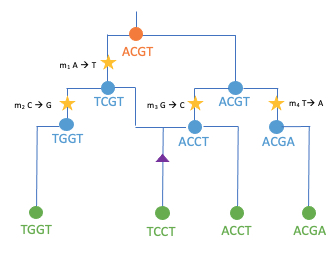
Fig.2: Schematic overviews of common ancestry. a.) Trees, describing how a collection of sets of chromosomes are related to each other at a chromosomal location. b.) Tree network, summary of a sequence of correlated trees when following the gene trees along a genome [inspired by [ 3, 4, 5]]
The aim of my work is to use new algorithms to derive all DNA segments in between from a collection of chromosome sets, a large number of maize individuals, the DNA segments of close relatives and the wild form of maize (teosinte), and thus be able to reconstruct the entire genealogy tree network for maize. Once I have done so, I will trace gene variants of interest within this family tree network, dating their first appearance and determining their geographic locations and historical migrations, and comparing these to the known history of maize. This would give us an unprecedented insight into the history and evolution of maize. All of this can help us understand how plants adapt to their environment. It would also show us which gene variants were particularly important for the adaptation of maize and allow us to reintroduce these gene variants into modern maize as well.
Planter's Punch
Under the heading Planter’s Punch we present each month one special aspect of the CEPLAS research programme. All contributions are prepared by our young researchers.
About the author

Kerstin Schulz is working on the domestication and colonization history of maize using ancestral recombination graphs. To do this, she tries to infer the age, origin and geographic distribution of all maize alleles in European maize. Her goal is to better understand the evolutionary history of different teosinte and maize populations along the maize expansion route and to identify the population genetic signals that shaped maize genetic diversity.
Picture references
[1] Chin Jian Yang et al. “The genetic architecture of teosinte catalyzed and constrained maize domestication”. In: Proceedings of the National Academy of Sciences 116.12 (2019), pp. 5643–5652.
[2] CIMMYT Maize Germplasm Bank. Maize: From Mexico to the world. 2016.