Prof. Dr. Paul Schulze-Lefert
Research focus
Structure and functions of the Arabidopsis rhizosphere microbiome
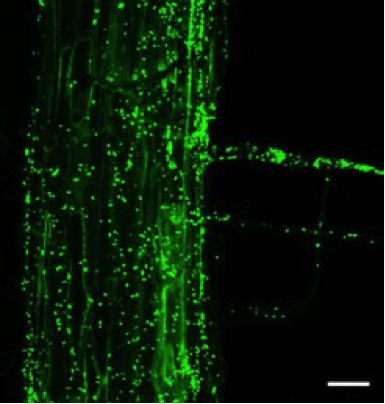
Remarkably little is understood about plant-microbe interactions that are, at first glance, ‘symptomless’. These poorly studied plant microbiomes harbor an unknown reservoir of probiotic and plant protective associations. One gram of soil typically contains ~108 to 1010 bacteria. Microbial DNA fingerprints from plant roots or aerial organs uncover microbial communities thriving on the surface of, or within, healthy plant tissue. Rhizosphere microbiomes attached to, and in the first few millimeters away from, the root surface are distinct from bulk soil, suggesting specific colonization events. Organic carbon flux from roots promotes the growth of microbial decomposers that, in turn, recycle plant nutrients for root uptake by transpiration-driven water fluxes. Seedlings exude 30-40%, and adult plants 20%, of photosynthetically fixed carbon into the rhizosphere in the form of poorly characterized rhizodeposits.
We have developed and applied bacterial 16S rRNA gene pyrosequencing to characterize and compare soil and root-inhabiting bacterial communities of Arabidopsis thaliana. We show that roots of Arabidopsis, grown in different natural soils under controlled environmental conditions, are preferentially colonized by Proteobacteria, Bacteroidetes and Actinobacteria, and each bacterial phylum is represented by a dominating class or family. Soil type defines the composition of root-inhabiting bacterial communities and host genotype determines their ribotype profiles to a limited extent. The identification of soil type-specific members within the root-inhabiting assemblies supports our conclusion that these represent soil-derived root endophytes. Surprisingly, plant cell wall features of other tested plant species appear to provide a sufficient cue for the assembly of ~40% of the Arabidopsis bacterial root-inhabiting microbiota, with a bias for Betaproteobacteria. Thus, this root sub-community may not be Arabidopsis-specific but saprophytic bacteria that would naturally be found on any plant root or plant debris in the tested soils. In contrast, colonization of Arabidopsis roots by members of the Actinobacteria depends on additional cues from metabolically active host cells (Bulgarelli et al., Nature 488, 91-95 (2012)).
The ten most important publications
- Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O'Connell RJ, Schulze-Lefert P (2016) Root Endophyte Colletotrichum tofieldiae Confers Plant Fitness Benefits that Are Phosphate Status Dependent. Cell 165(2):464-474.
- Hacquard S, Kracher B, Hiruma K, Münch PC, Garrido-Oter R, Thon MR, Weimann A, Damm U, Dallery JF, Hainaut M, Henrissat B, Lespinet O, Sacristán S, Ver Loren van Themaat E, Kemen E, McHardy AC, Schulze-Lefert P, O'Connell RJ (2016) Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat Commun 7:11362.
- Dombrowski N, Schlaeppi K, Agler MT, Hacquard S, Kemen E, Garrido-Oter R, Wunder J, Coupland G, Schulze-Lefert P (2016) Root microbiota dynamics of perennial Arabis alpina are dependent on soil residence time but independent of flowering time. ISME J.
- Hacquard S, Garrido-Oter R, Gonzalez A, Spaepen S, Ackermann G, Lebeis S, McHardy AC, Dangl JL, Knight R, Ley R, Schulze-Lefert P (2015) Microbiota and Host Nutrition across Plant and Animal Kingdoms. Cell host & microbe 17(5):603-616.
- Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell host & microbe 17(3):392-403.
- Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, Hüttel B, McHardy AC, Vorholt JA, Schulze-Lefert P (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528(7582):364-369.
- Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P (2014) Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci U S A 111(2):585-592.
- Guttman DS, McHardy AC, Schulze-Lefert P (2014) Microbial genome-enabled insights into plant-microorganism interactions. Nature reviews. Genetics 15(12):797-813.
- Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annual review of plant biology 64:807-838.
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488(7409):91-95.
Prof. Dr. Paul Schulze-Lefert

Department of Plant Microbe Interactions
Max Planck Institute for Plant Breeding Research