Prof. Dr. Oliver Ebenhöh
Research focus
My research group develops mathematical models of plant energy metabolism and photosynthesis. One focus lies on the investigation of acclimation processes of the electron transport chain: how is the trade-off between maximal photosynthetic yield and protection against damaging light intensities achieved? We also develop novel methods to simulate and describe the production and degradation of biopolymers such as starch and complex secondary metabolites. Thirdly, we are interested in understanding the timing of metabolism: how are key processes regulated by the diurnal rhythm and the circadian clock?
The ten most important publications
- Pokhilko A, Ebenhöh O (2015) Mathematical modelling of diurnal regulation of carbohydrate allocation by osmo-related processes in plants. Journal of the Royal Society, Interface / the Royal Society 12(104):20141357.
- Pokhilko A, Bou-Torrent J, Pulido P, Rodríguez-Concepción M, Ebenhöh O (2015) Mathematical modelling of the diurnal regulation of the MEP pathway in Arabidopsis. New Phytol 206(3):1075-1085.
- Pokhilko A, Flis A, Sulpice R, Stitt M, Ebenhöh O (2014) Adjustment of carbon fluxes to light conditions regulates the daily turnover of starch in plants: a computational model. Molecular bioSystems 10(3):613-627.
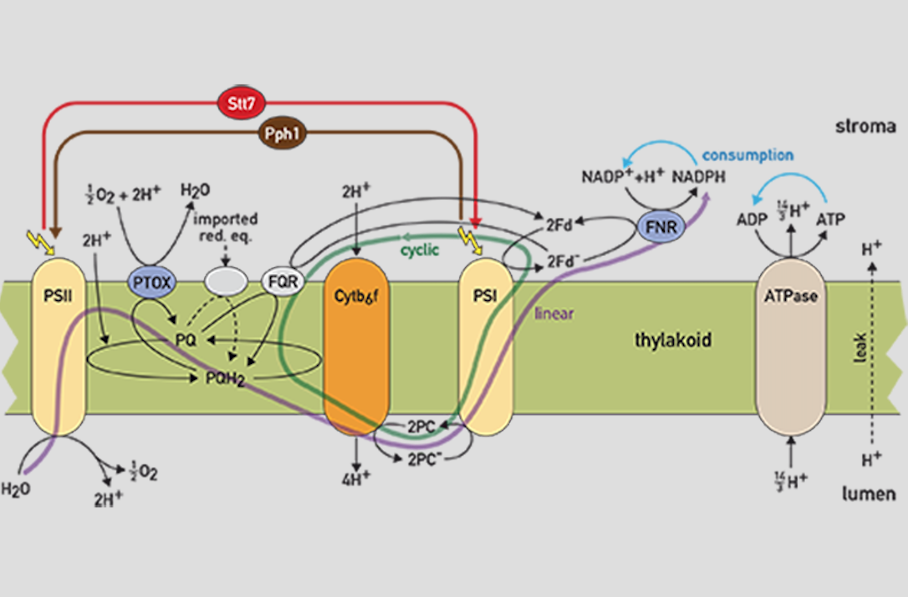
- Ebenhöh O, Fucile G, Finazzi G, Rochaix JD, Goldschmidt-Clermont M (2014) Short-term acclimation of the photosynthetic electron transfer chain to changing light: a mathematical model. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 369(1640):20130223.
- Kartal O, Ebenhöh O (2013) A generic rate law for surface-active enzymes. FEBS letters 587(17):2882-2890.
- Kartal O, Mahlow S, Skupin A, Ebenhöh O (2011) Carbohydrate-active enzymes exemplify entropic principles in metabolism. Mol Syst Biol 7:542.
- Witzel F, Götze J, Ebenhöh O (2010) Slow deactivation of ribulose 1,5-bisphosphate carboxylase/oxygenase elucidated by mathematical models. FEBS J 277(4):931-950.
- May P, Wienkoop S, Kempa S, Usadel B, Christian N, Rupprecht J, Weiss J, Recuenco-Munoz L, Ebenhöh O, Weckwerth W, Walther D (2008) Metabolomics- and proteomics-assisted genome annotation and analysis of the draft metabolic network of Chlamydomonas reinhardtii. Genetics 179(1):157-166.
- Handorf T, Christian N, Ebenhöh O, Kahn D (2008) An environmental perspective on metabolism. Journal of theoretical biology 252(3):530-537.
- Matuszynska A (BBA Bioenergetics (in press).
Prof. Dr. Oliver Ebenhöh

Institute for Quantitative and Theoretical Biology
Heinrich Heine University Düsseldorf