Dr. Shishir K Gupta
Research focus
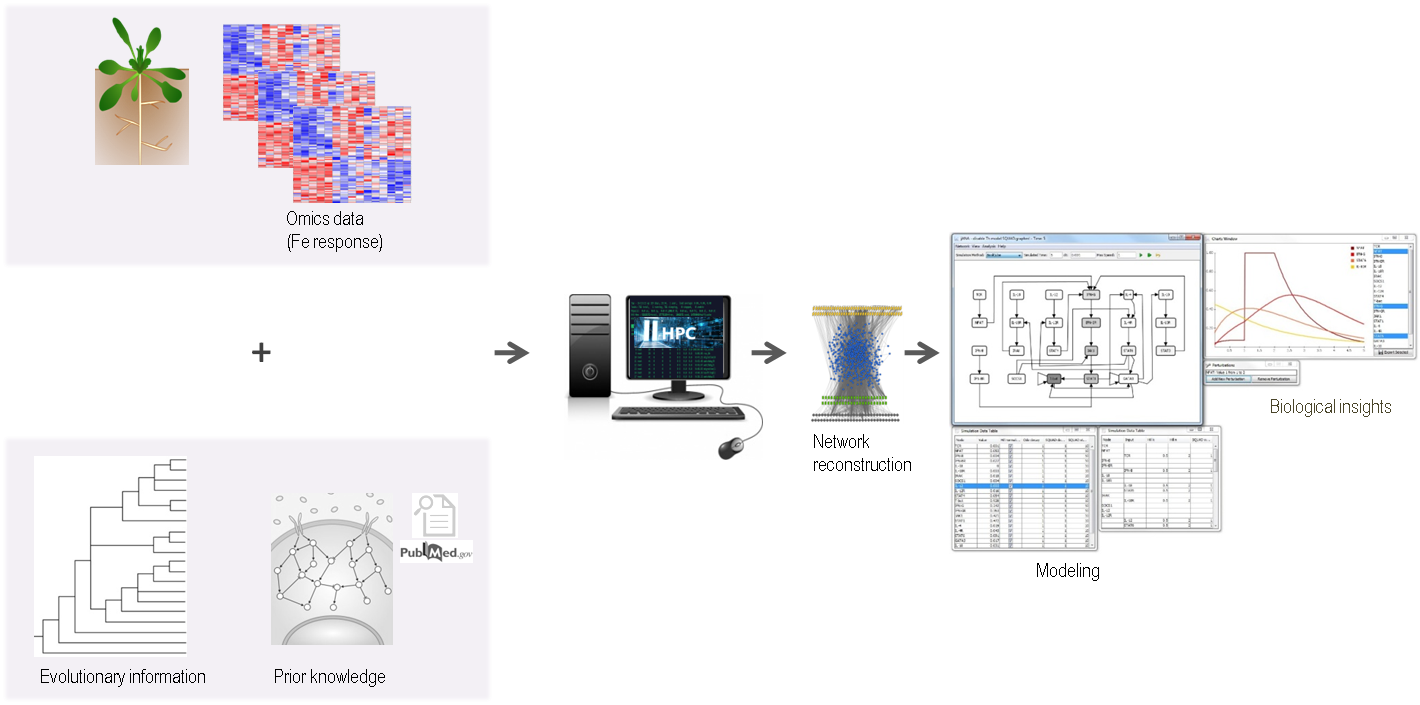
Iron is an essential micronutrient for plants. It is vital for the growth and health of the plant. At the same time, iron is very toxic when it accumulates to high levels. As a result, plants closely regulate Fe homeostasis and respond to both Fe overload and deficiency. In the last decades, several transporters and regulatory factors involved in this process have been isolated and identified. However, it is unclear how the known and undiscovered components of Fe regulation operate together as a regulatory network. My focus is to reconstruct a mechanistic and predictive model of Fe regulation to improve our understanding of the molecular network that underlies Fe homeostasis. To achieve this goal, I am performing an analysis of large-scale gene expression data collected from mutant studies and integrating it with the prior knowledge network of Fe regulation. Evolutionary information from other plants will also be used to identify the potential regulatory network components. Model simulations and analysis will be performed with particular focus on the coherent and incoherent regulation of sub-networks in Fe deficiency response regulation. The model will not only simulate the Fe response, but will also predict the involvement of additional transcription factors and proteins not previously associated with Fe homeostasis.
Computational modelling of Fe-regulatory networks
Cellular signalling can be amplified in positive feed-forward loops or become inhibited under a negative feed-back. Nutritional cues often elicit complex configurations of coherent and incoherent regulation. The micronutrient iron (Fe) is required for many redox and electron transfer reactions. Fe affects photosynthesis and nutrient assimilation and determines plant nutritional values. During their growth, plants are challenged to take up Fe from the soil, store it, transport and allocate it to sink organs, in accordance with growth dynamics and environmental stress as experienced under climate change. An intricate gene regulatory network controls Fe homeostasis. The aim of this project is to combine large-scale data to reconstruct mechanistic models and simulate the Fe-regulatory response networks. Regulatory circuits are experimentally validated in targeted assays.
The five most important publications
- Gupta, S.K., Srivastava, M., Osmanoglu, Ö., Xu, Z., Brakhage, A.A. and Dandekar, T., 2021. Aspergillus fumigatus versus Genus Aspergillus: Conservation, adaptive evolution and specific virulence genes. Microorganisms, 9(10), p.2014.
- Akhoon, B.A., Gupta, S.K. and Dhar, M.K., 2023. Dissecting the genome, secretome, and effectome repertoires of Monilinia spp.: The causal agent of brown rot disease: A comparative analysis. Postharvest Biology and Technology, 195, p.112120.
- Gupta, S.K., Ponte-Sucre, A., Bencurova, E. and Dandekar, T., 2021. An Ebola, Neisseria and Trypanosoma human protein interaction census reveals a conserved human protein cluster targeted by various human pathogens. Computational and structural biotechnology journal, 19, pp.5292-5308.
- Gupta, S.K., Srivastava, M., Osmanoglu, Ö. and Dandekar, T., 2020. Genome-wide inference of the Camponotus floridanus protein-protein interaction network using homologous mapping and interacting domain profile pairs. Scientific reports, 10(1), pp.1-12.
- Srivastava, M., Bencurova, E., Gupta, S.K., Weiss, E., Löffler, J. and Dandekar, T., 2019. Aspergillus fumigatus challenged by human dendritic cells: metabolic and regulatory pathway responses testify a tight battle. Frontiers in cellular and infection microbiology, 9, p.168.
