Family Business
A gene family is a collection of genes which originated from a single gene though gene or genome duplication events, which have similar nucleotide or protein sequences, and perform usually similar functions. This, or a somewhat similar phrase, is the definition every biology student learned, hopefully, during the curriculum. In the field of biology, this is a pretty straight forward definition, considering the jury is still out on defining the “gene”.
The attentive reader will have guessed the overall topic of this Planter Punch issue. Further, in this issue a “gene” will describe the continuous DNA sequence between the beginning and and end of a gene. And finally, let me explain why analyzing plant gene families matters. Analyzing and comparing plant gene families solves basic scientific questions, e.g., how plants diversify and supports practical tasks, e.g., asses the completeness of a sequenced plant genome. Genome sequencing is the process of decoding and determining the DNA sequence of an organism. Plants diversify by accumulating evolutionary changes within or between plant species. These changes may improve the survivability of a plant species and allow it to thrive in a different or changing environment, e.g., due to climate change or by gaining a resistance to a plant pathogen.
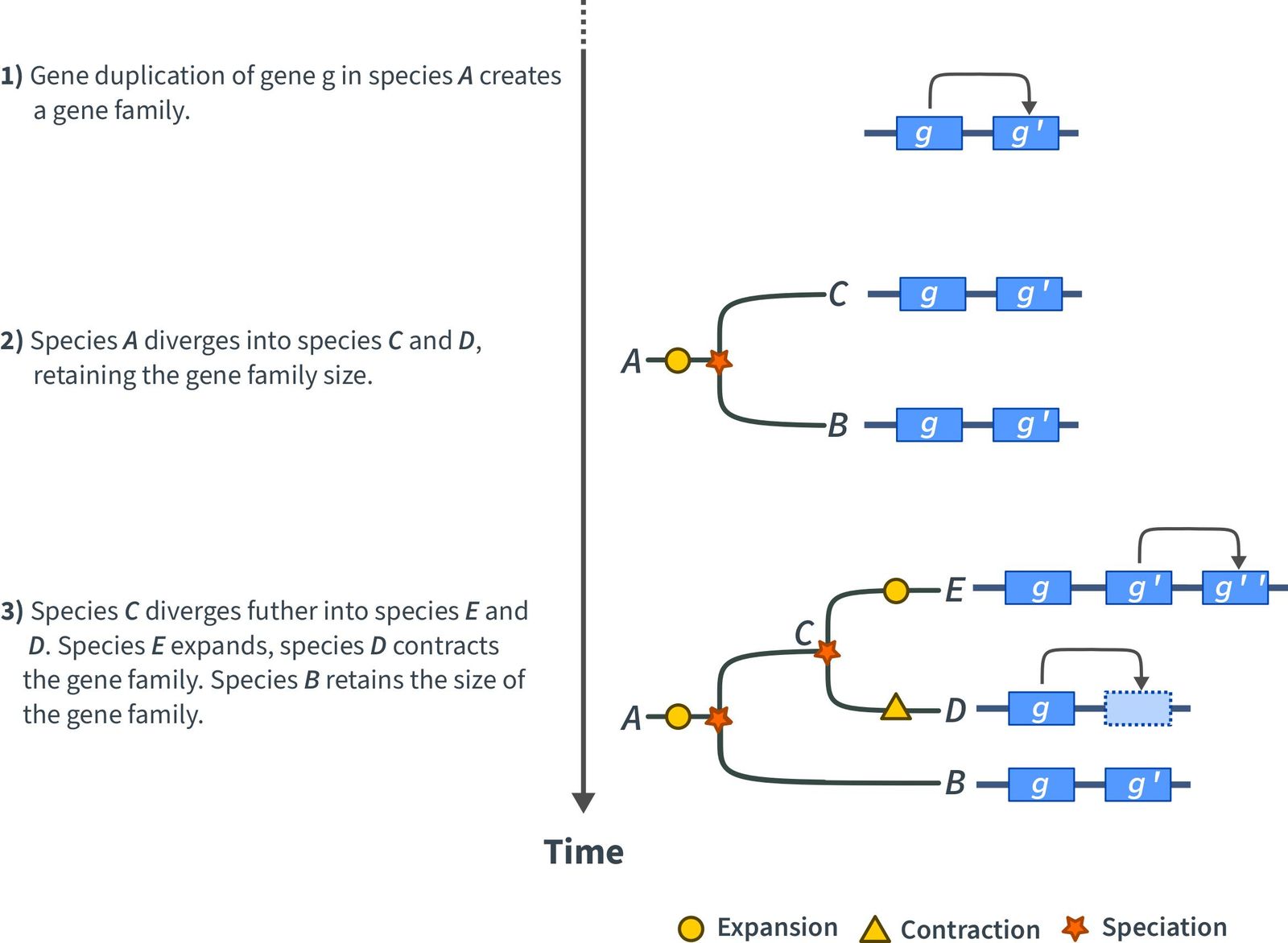
The duplication, or loss, of a gene can have a strong impact onto the fitness and evolution of a plant. The impact of a duplication or deletion depends on the importance of the gene in question and its role in the pathways it is, or was, involved. If a gene is duplicated, the gene family expands, i.e., it grows. If a gene is lost, the gene family contracts, i.e., it shrinks (Figure 1). The size of a gene family varies and can range from two members up to more than hundred. In addition, the size of a gene family between two species can vary as well. However, we assume that the size of a gene family involved in basic and important pathways are very similar.
Expansion or contraction of individual gene families within a plant lineage occurs randomly or is the result of natural selection, i.e., a change in a gene family did benefit a plant in some way. Analyzing gene families helps to identify such events and the subsequent influence on the plant evolution. Including more diverged plant species in such studies, that is, plants which are evolutionary more distant, e.g., Arabidopsis thaliana, Triticum aestivum (bread wheat), mosses, algae, can help to pinpoint when the changes did occur (Figure).
Before any comparison can be performed, gene families must be defined and identified. Both steps are usually performed using protein sequences. Proteins are individual amino acids linked together to form a chain. A gene encodes these individual amino using a specific codon for each amino acid and the order of these codons within a gene encode a protein. It is possible that a single amino acid can be encode by several codons, e.g., different codons can result in the same amino acid. This is called the codon degeneracy, or redundancy, of the genetic code. Thus, proteinsequences between different species may be more similar than the underlying genetic code. This facilitates identifying sequences with similar functions from more diverged sequences.
To define a gene family, known proteins with similar sequences are grouped together to create protein clusters, so that each cluster can be assigned a biological function. Eventually, each specific cluster encoded in a model which can be used to screen existing or new genomes for specific gene family members. The individual models are manually curated and continuously updated. Further, it allows to identify gene families very fast. The proteome, i.e., all known proteins, of one plant species can be analyzed in matter of minutes.
The current, and steady increasing, abundance of available genome data allows us now to perform comparisons between scores of plant species, some being very different. To this end, we are applying high throughput approaches to screen the proteomes of these plants to identify expansions and contractions within known gene families and to identify species specific gene families. In addition, we are trying to identify when these important events occurred during the evolution of different plant species.
Protein sequences facilitate the identification and comparison of gene families because i) they facilitate the comparison of diverge plant species; ii) they are available in abundant quantities; iii) tools and methods to handle this data are available and thoroughly tested. Proteins are not linear constructs but fold into characteristic structures and individual structures can interact to form larger and more complex structures. These structures determine the function of a protein, for example an enzyme. Remarkably, very different protein sequences can result in similar structures performing similar functions.
The question arises why are we then still using protein sequences to identify gene families? A main obstacle so far is obtaining protein structures because this process is experimentally still complex and lengthy. However, scientists and engineers from academia and private companies have made recently big advances in developing software to predict protein structures. Unfortunately, theseapproaches still require huge amounts of computer power and hardware. Nonetheless, this will not be the silver bullet in analyzing gene families. Similar protein structures between species can either originate from a single gene in an ancestral species or evolved independently between species. This process is called “convergent evolution”. Examples of convergent evolution in plants is the C4 photosynthesis or carnivorous plants. These abilities did not evolve once in an ancestor species but evolved several times within the specific plant families. Another example is the evolution of the eye within vertebrates (including humans), Cephalopods (e.g., kraken), and Cnidaria (e.g., jellyfish), which looks and functions very similar but did evolve independently within each of these groups. Convergent evolution must be included when comparing protein structures, the analysis of gene families being an appropriate tool.
With protein sequences we will not identify possible gene family members from very diverged species, but convergent evolution has a lower impact on our analyses. On the other hand, protein structures would allow us to find putative gene family members form very different species and different evolutionary pathways will have a stringer influence on our analyses. Yet, before we can analyze gene families in plants using protein structures, more structures need to be solved experimentally, or predict and tested. Until then, we will rely on protein sequences in our analyses.
Planter’s Punch
Under the heading Planter’s Punch we present each month one special aspect of the CEPLAS research programme. All contributions are prepared by our early career researchers.

About the author
Jan Buchmann is postdoctoral researcher in the group of Prof. Björn Usadel in the Institute for Biological Data Science at the Heinrich-Heine-University. His research focuses on the evolution of gene families in plants and developing methods to identify plant gene sub-families in genomes without prior gene calling. He earned his PhD at the University of Zurich in Switzerland analyzing the influence of transposons on plant genome evolution. Before he joined CEPLAS, he researched virus genome evolution and developed methods to identify novel viruses in metagenomic data sets at the University of Sydney in Australia.