Forward genetics – A powerful tool to find novel genes responsible for C4 leaf anatomy
Plants using C4 photosynthetic pathway exhibit higher CO2, nitrogen and water use efficiencies than C3 plants and thereby they can grow better under high light intensities, high temperatures and dryness. One of the major reasons for their adaptive advantage is the evolution of the C4 specific Kranz leaf anatomy. Plants with Kranz anatomy have high number of veins close to each other and surrounded by enlarged bundle-sheath cells, loaded with chloroplasts. But the genetic regulatory network responsible for the development of Kranz anatomy is largely unknown.
Forward genetics is a large scale screening approach to determine the genetic basis responsible for a phenotype. The basis of this screening method is creating random mutations in the genome. We are using two forward genetic approaches to screen for mutant phenotypes with C4 specific Kranz anatomy and to discover responsible genes. First one is Activation tagging and second one is EMS mutagenesis. Activation tagging is nothing but to over-express the nearby genes by randomly inserting T-DNA region harbouring a strong promoter. Whereas in EMS mutagenesis, the alkylating agent EMS (ethyl methanesulfonate) is used to induce single base pair changes in the genome that often result in creating loss-of-function mutants.
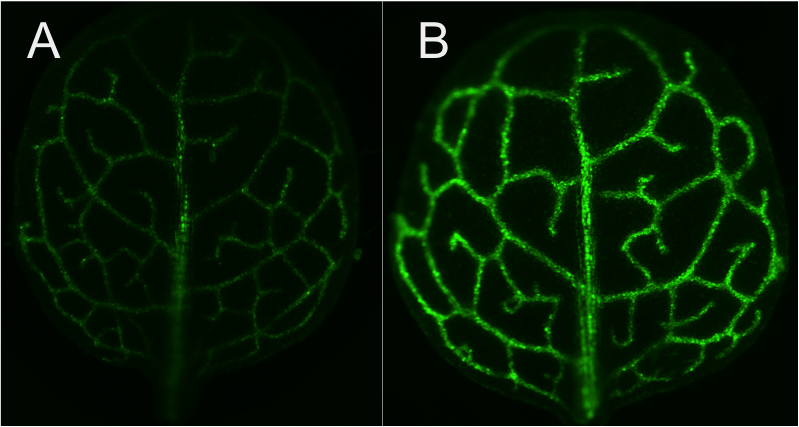
For our screenings, we are using Arabidopsis thaliana, a well-known model species for plant scientists, which is a C3 plant and has basic blueprint of Kranz anatomy, small bundle sheath cells with few chloroplasts. To easily identify plants with changed leaf anatomy, Arabidopsis thaliana bundle-sheath cells are labelled with chloroplast targeted Green Fluorescent Protein (GFP) and this GFP marker line has been subjected to both Activation tagging and EMS mutagenesis. Since each chloroplast in the bundle sheath cells is labelled with GFP, we assume that GFP signal intensity should correlate with chloroplast number and thus with bundle sheath cell size. So we detached single leaf from each plant and observed at the microscope with GFP filter to select plants with more or less GFP signal intensity, later we verify the bundle-sheath cell chloroplast number and cell size and isolate candidate genes using inverse PCR and mapping by sequencing methods.
The identified interesting candidates will be further analysed in crop plants like rice.
The major crop plants like rice, wheat, barley and soybean are C3 plants. To improve their photosynthetic efficiency under changing climatic conditions, we need to engineer C4 photosynthetic pathway, basic understanding of Kranz anatomy development is prerequisite for this.
Contribution by Kumari Billakurthi, Developmental and Molecular Biology of Plants, HHU
Planter’s Punch
Under the heading Planter’s Punch we present each month one special aspect of the CEPLAS research programme. All contributions are prepared by our young researchers.
Corresponding publication
Gowik U, Westhoff P (2011) The Path from C3 to C4 Photosynthesis. Plant physiology 155(1):56-63. [Abstract]