Bacteria – diverse applications as microbial cell factories
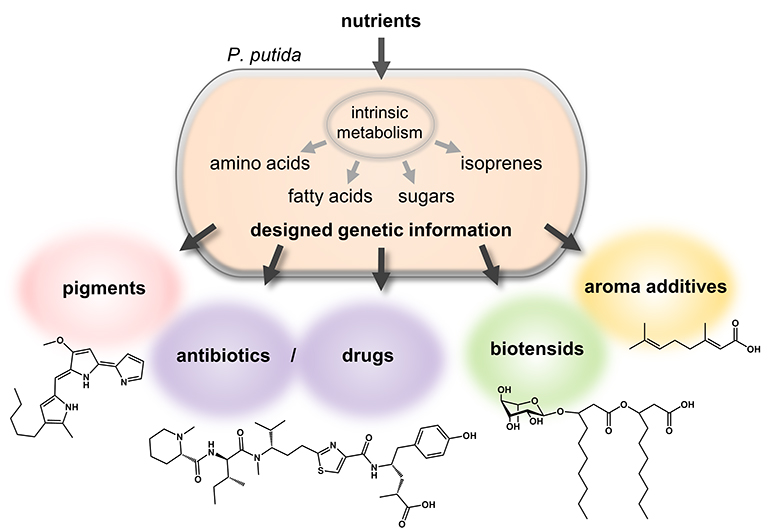
Bacteria are able to take up simple molecules like sugars or alcohols as nutrients and utilize them to build all components that are necessary for basic cell metabolism, thus so called primary metabolites, via their intrinsic enzyme machinery. In addition, like plants bacteria can synthesize a diverse spectrum of often species-specific secondary metabolites. These are more complex molecules which fulfill distinct functions beyond the primary metabolism. Because of these specific functions, e.g. repelling of competing organisms, they are often of great value, e.g. as antibiotics for human use. Since the recovery of such secondary metabolite-based high-valuable compounds from natural producers is typically limited, there is an increasing demand to establish especially suited bacteria as designer cell factories for this purpose.
Via the targeted transfer of DNA carrying genetic information for the biosynthesis of valuable secondary metabolites in diverse microorganisms, like Escherichia coli, Rhodobacter capsulatus and P_seudomonas putida_, many such metabolites could already be synthesized (Figure 1, Liebl et al., 2014; Loeschcke & Thies 2015). One example is the biosynthesis of the antimicrobial pigment prodigiosin in P. putida (Loeschcke et al., 2013, Domröse et al., 2015).
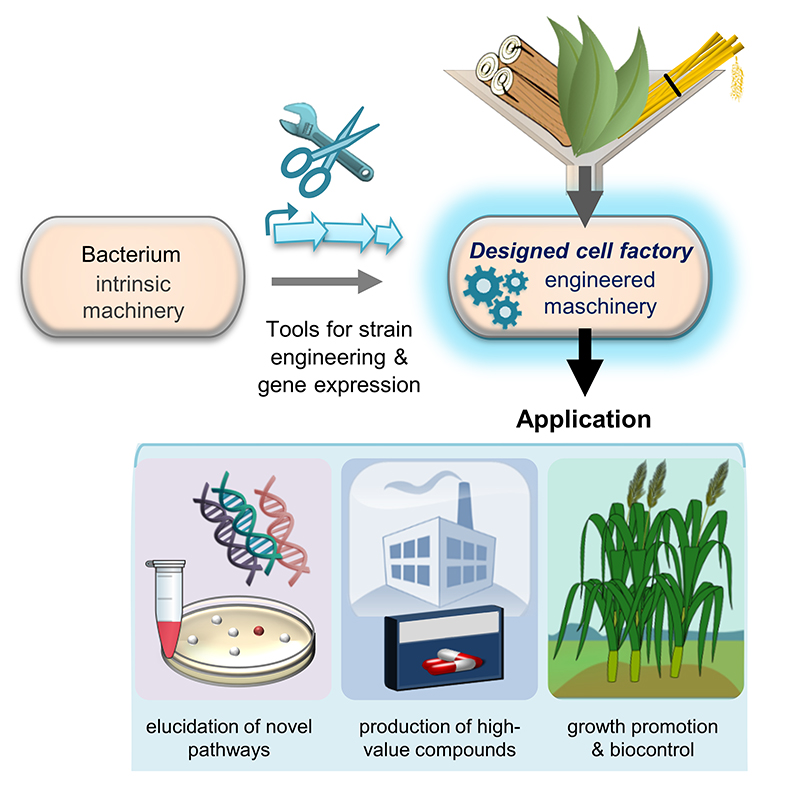
Likewise constructed microbial cell factories have a broad application potential within CEPLAS research areas C and D (Figure 2). They enable the elucidation and investigation of new bacterial and plant secondary metabolite pathways, as well as the high-level production of the respective compounds allowing their characterization and application. Potential applications exist e.g. in the field of sustainable plant cultivation, since several metabolites are known to improve plant health and growth. This can be effectuated via different mechanisms. Certain metabolites can, for instance, directly trigger enhanced plant growth as signal molecules; others can indirectly support plant growth by inhibition of plant pathogens or by shaping a beneficial microbiome around the plant.
Contribution by Anita Loeschcke, Institute of Molecular Enzyme Technology , HHU/FZJ
Planter’s Punch
Under the heading Planter’s Punch we present each month one special aspect of the CEPLAS research programme. All contributions are prepared by our young researchers.
Corresponding publications
Loeschcke A, Thies S (2015) Pseudomonas putida-a versatile host for the production of natural products. Applied microbiology and biotechnology 99(15):6197-6214. [Abstract]
Domrose A, Klein AS, Hage-Hulsmann J, Thies S, Svensson V, Classen T, Pietruszka J, Jaeger KE, Drepper T, Loeschcke A (2015) Efficient recombinant production of prodigiosin in Pseudomonas putida. Frontiers in microbiology 6:972. [Abstract]
Liebl W, Angelov A, Juergensen J, Chow J, Loeschcke A, Drepper T, Classen T, Pietruszka J, Ehrenreich A, Streit WR, Jaeger KE (2014) Alternative hosts for functional (meta)genome analysis. Applied microbiology and biotechnology 98(19):8099-8109. [Abstract]
Loeschcke A, Markert A, Wilhelm S, Wirtz A, Rosenau F, Jaeger KE, Drepper T (2013) TREX: a universal tool for the transfer and expression of biosynthetic pathways in bacteria. ACS synthetic biology 2(1):22-33. [Abstract]