Insight into the regulation of flowering
Plants from the same species can occur with different phenotypes, e.g. some plants have more leaves or flower earlier than others. This diversity can be due to variations in gene transcription, which is controlled by transcription factors. Transcription factors are proteins that switch genes on or off and thereby can regulate the expression of proteins.
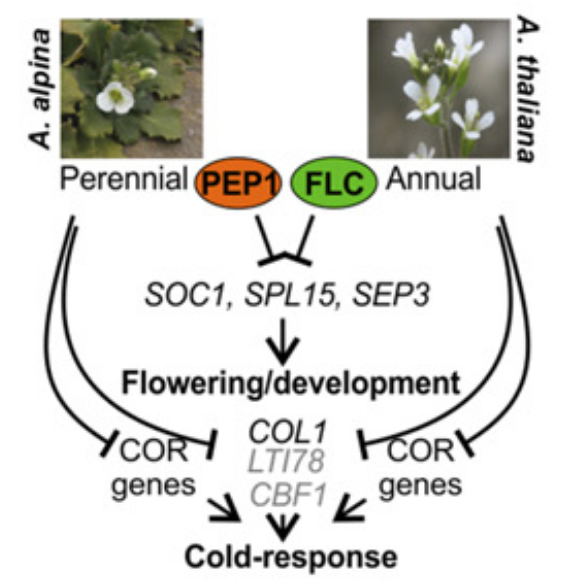
The complex process of flowering in higher plants is one example, where a transcription factor plays a major role in the regulation of plant development. In the model plant Arabidopsis thaliana, the transcription factor FLOWERING LOCUS C (FLC) represses the development of flowers until plants are exposed to cold in wintertime. However, it’s not well understood yet, how transcriptional patterns evolved. The analysis of transcriptional patterns has been addressed recently at the genome-wide level in yeast and metazoans (ChIPseq experiments) and as well in plants from the same species, but rarely between plants from different species. This question has now been addressed in the new paper <link https: www.ncbi.nlm.nih.gov pubmed _blank external-link-new-window internal link in current>“Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species” with contribution from CEPLAS Research Area A coordinator <link _blank internal-link internal link in current>George Coupland and CEPLAS Alumni <link _blank internal-link internal link in current>Vicky Tilmes.
In the current study, target genes (binding sites) of the A. thaliana transcription factor FLC were compared with target genes from the species Arabis alpina, which is as well from the plant family Brassicaceae. Therefore, the authors first had to determine the target genes of the FLC counterpart in A. alpina, which is the orthologous transcriptional factor PERPETUAL FLOWERING 1 (PEP1). Both transcriptional factors (FLC and PEP1) are known as repressors of flowering. Then, target genes were compared between both species. Interestingly less than 20% of the target genes were conserved across species; however, many of those conserved genes were involved in flowering. The comparison of target genes across species therefore seems a promising approach to identify which genes are involved in the core function of a transcription factor; which is in this case floral induction.
The remaining 80% of analysed target genes were different between the annual plant A. thaliana and the perennial plant A. alpina and were often involved in stress responses. This high difference of target genes of orthologous transcription factors suggests that transcription factor binding sites in plants change rapidly during evolution. Most likely this is due to the fact that in both analysed plants the species specific target genes are involved in environmental stress responses and have adapted to environmental changes and different habitats. This suggests that those binding sites evolved later than the ones involved in flowering.
Transcription factors control important traits like flowering. In this study, for the first time two plant transcription factors from different species were compared and it was shown that the the core function (flowering) is conserved while other functions (environmental stress) differ between species.
Full publication: Mateos JL, Tilmes V, Madrigal P, Severing E, Richter R, Rijkenberg CWM, Krajewski P, Coupland G (2017) Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. Proc Natl Acad Sci U S A. 114(51):E11037-E11046. doi: 10.1073/pnas.1618075114 <link https: www.ncbi.nlm.nih.gov pubmed _blank external-link-new-window internal link in current>[Abstract]
<link _blank internal-link internal link in current>Learn more about CEPLAS Research Area A